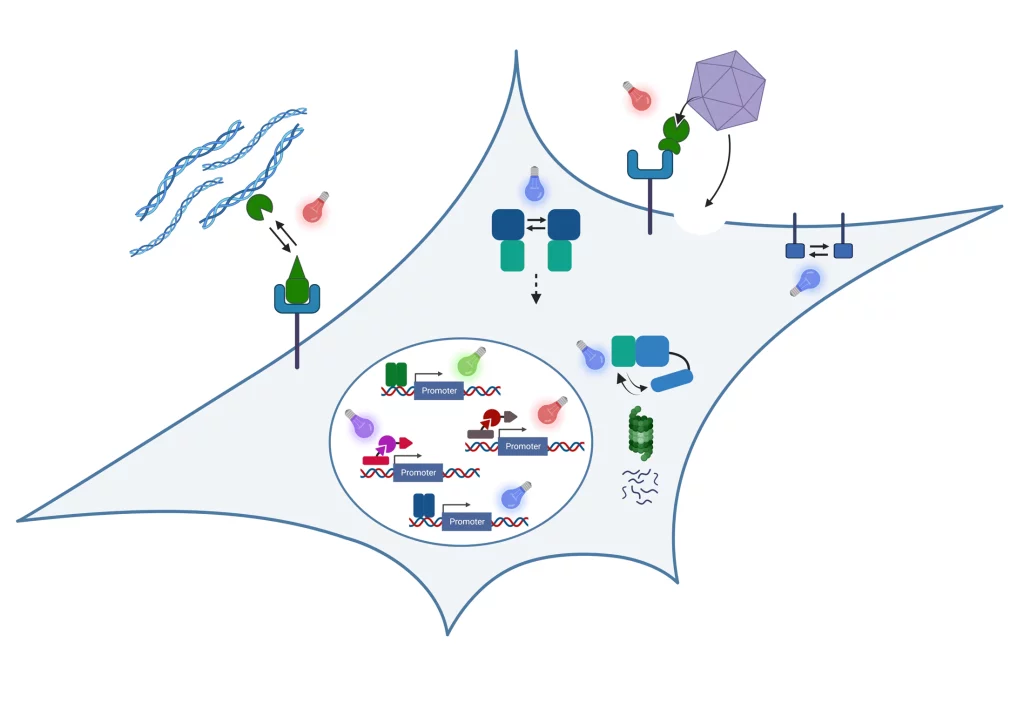
Our inspiration is the ability of organisms and the materials they are made of to adapt to dynamic environmental conditions. Plants adapt growth to light conditions; bacteria develop resistance against antibiotics or bones get stronger when exercised. The basis for this ability to adapt is a fascinating information processing machinery of the organisms: Environmental conditions are captured by molecular sensors, then the signals are processed and integrated with genetic programs to finally yield a targeted response.
In our research, we engineer nature’s molecular sensing, processing, and actuation machinery in order to precisely control the function and properties of cells and materials. We apply these newly developed technologies in different fields of fundamental and applied research.

Team Members
Job Openings
Looking for a fascinating position at the intersection of synthetic biology and materials sciences as PhD student, postdoc, or visiting scientist? We welcome your spontaneous application. To get a glimpse of how we work together in our department, please check out the following video
Research
Stimulus-responsive and Information-processing (living) Materials
We develop and apply stimulus-responsive and information-processing biohybrid polymer materials. To this aim, we functionally couple synthetic biological molecular sensors and switches to polymer materials. By wiring these switches according to topologies inspired by electronic circuits, we engineer materials that perform fundamental computational operations. Examples of our work include:
- We engineered a hydrogel based on a bacteria-derived photoreceptor which allows the light-responsive, fully reversibly tuning of its mechanical properties. We applied this hydrogel as extracellular matrix to analyze the impact of dynamic mechanical environments on transcriptome-wide responses in mesenchymal stem cells or on the migration of T-lymphocytes.
See Hörner et al. Advanced Materials 2019 - We integrated synthetic biological switches with polymer materials into a circuit inspired by an electronic counter. The resulting material system was able to count the number of input light pulses and to release different output as a function of the number of light pulses detected. We applied this system to sequentially release different biocatalysts to drive a two-step biochemical reaction.
See Beyer et al., Advanced Materials 2018 - We developed PenTag, a protein tag for the spontaneous, covalent coupling of proteins to ampicillin-functionalized molecules such as dyes, polymers, or solid supports. Based on this strategy, we engineered and assembled material modules to function as encoder for processing different combinations of biochemical input stimuli.
See Mohsenin et al., Advanced Functional Materials 2024 - By engineering modular protease-based switches that can either be activated or repressed, we develop information-processing biohybrid circuits that process binary biomolecular information according to a circuit inspired by electronic decoders. Such circuits can be applied to process and interpret biochemical sensor information for advanced diagnostic applications.
See Mohsenin et al., Advanced Materials 2024
Molecular optogenetics to control cell fate and function
We develop and apply molecular optogenetic tools to control cell fate and function with unprecedented spatial and temporal precision in a dose-dependent and highly specific manner. To this aim, we engineer plant- and bacteria-derived photoreceptors and functionally couple them to proteins involved in cell signaling and gene expression. Examples of our work include:
- Light-inducible formation of liquid or gel-like transcription factor condensates in mammalian cells and mice. We demonstrate that liquid “transcription factor droplets” show a several-fold higher activity in inducing transgene expression compared to native transcription factors. Further, gel-like transcription factor condensates were shown to correlate with decreased transcriptional activation thus providing a materials-based layer of controlling gene expression.
See Schneider et al., Science Advances 2021 and Fischer et al., Small 2024 - Light-guided adeno-associated viral (AAV) vectors. We engineered a light-responsive tropism into AAVs which allows us to selectively transfer genetic information into single cells or to transduce different cells within one culture with different transgenes.
See Hörner et al., Science Advances 2021
Our group is running www.optobase.org, the most comprehensive database on molecular optogenetics. Have a look and discover the amazing opportunities in controlling biology with light!
Biosensors
We integrate natural and engineered molecular sensors for drugs, metabolites or nucleic acids into suitable readout formats for the fast and sensitive quantification of such substances. Together with collaboration partners, we develop biosensor systems for different application fields:
Projects and Partners
We perform collaborative research in materials-oriented synthetic biology within interdisciplinary research consortia
STEADY
Within the ERC Advanced Grant STEADY, we develop concepts for dynamically controlling the properties of engineered living materials by advanced synthetic genetic circuits.
LoopOfFun
We coordinate the European Innovation Council (EIC)-funded consortium LoopOfFun in which we aim at developing a platform for the rapid development of industry-scale, one-step, simple casting-based manufacturing processes for fungal mycelia composites. We jointly work towards this goal with our consortium partners:
- Prof. Roman Jerala, National Institute of Chemistry, Ljubljana, Slovenia
- Dr. Achim Weber, Fraunhofer IGB, Stuttgart, Germany
- Prof. Arnold Driessen, University of Groningen, The Netherlands
- Carlotta Borgato and Jan Boelen, Atelier LUMA, Arles, France
DELIVER
In the project DELIVER funded by the Carl-Zeiss-Foundation, we collaborate towards the data-driven engineering of sustainable living materials. We combine synthetic biology with materials sciences and data-driven approaches to design bio-based composite materials with custom-tailored structural properties for construction applications. Within deliver, we collaborate with the following partners:
- Prof. Thomas Speck, University of Freiburg, Germany
- Dr. Clemens Kreutz, University Hospital Freiburg, Germany
BILLARD
We coordinate the BILLARD project funded by the Federal Ministry of Education and Research (BMBF) within the funding line “Biologization of Technology”, we collaborate with PD Dr. Felicitas Bucher from the Clinic of Ophtamology at the University Hospital Freiburg on the development of novel intraocular drug delivery devices.
CIBSS – Centre for Integrative Biological Signalling Studies
We are member of the Cluster of Excellence CIBSS in which we perform research on novel optogenetic technologies to control signaling reactions in mammalian cells. We mainly collaborate with Prof. Dr. Jens Timmer on the model-based design of synthetic biological switches and networks and with Prof. Dr. Wolfgang Schamel on controlling immunological processes such as T cell activation via optogenetics.
Publications
Malphettes, L. | Weber, C. C. | El-Baba, M. D. | Schoenmakers, R. G. | Aubel, D. | Weber, Wilfried | Fussenegger, M.
Nucleic Acids Research , 2005, 33 (12), 1-13.
https://dx.doi.org/10.1093/nar/gni107
Weber, C. C. | Cai, H. | Ehrbar, M. | Kubota, H. | Martiny-Baron, G. | Weber, Wilfried | Djonov, V. | Weber, E. | Mallik, A. S. | Fussenegger, M. | Frei, K. | Hubbell, J. A. | Zisch, A. H.
Journal of Biological Chemistry , 2005, 280 (23), 22445-22453.
https://dx.doi.org/10.1074/jbc.M410367200
Weber, C. C. | Link, N. | Fux, C. | Zisch, A. H. | Weber, Wilfried | Fussenegger, M.
Biotechnology and Bioengineering , 2005, 89 (1), 9-17.
https://dx.doi.org/10.1002/bit.20224
Weber, Wilfried | Malphettes, L. | de Jesus, M. | Schoenmakers, R. | El-Baba, M. D. | Spielmann, M. | Keller, B. | Weber, C. C. | van de Wetering, P. | Aubel, D. | Wurm, F. M. | Fussenegger, M.
Journal of Gene Medicine , 2005, 7 (4), 518-525.
https://dx.doi.org/10.1002/jgm.682
Weber, Wilfried | Malphettes, L. | Rinderknecht, M. | Schoenmakers, R. G. | Spielmann, M. | Keller, B. | Van De Wetering, P. | Weber, C. C. | Fussenegger, M.
Biotechnology Progress , 2005, 21 (1), 178-185.
https://dx.doi.org/10.1021/bp0498995
Weber, Wilfried | Rimann, M. | De Glutz, F. N. | Weber, E. | Memmert, K. | Fussenegger, M.
Metabolic Engineering , 2005, 7 (3), 174-181.
https://dx.doi.org/10.1016/j.ymben.2005.01.003
Weber, Wilfried | Spielmann, M. | El-Baba, M. D. | Keller, B. | Aubel, D. | Fussenegger, M.
Biotechnology and Bioengineering , 2005, 90 (7), 893-897.
https://dx.doi.org/10.1002/bit.20482